Multi-omic analysis to identify risk signatures in histologically normal breast tissue to stratify screening and prevention strategies
Primary supervisor: J Louise Jones, Queen Mary University of London
Secondary supervisor: Anita Grigoriadis, King’s College London
Project
Breast cancer is the commonest cause of cancer-related deaths among women worldwide. Women with germline (GL) BRCA1/2 mutations or previous history of breast cancer have a higher risk of developing malignancy, however, this risk varies between individuals. Our current challenge is to provide more precise risk estimates in order to stratify screening and preventive strategies. To do this, we require an in-depth understanding of physiological processes underlying tumour initiation in the normal breast tissue, to establish biomarkers which robustly identify truly high-risk individuals.
There is evidence that genomic alterations precede the morphological features of malignancy. Normal breast tissue from risk-reducing mastectomies (RRM) in women with GL BRCA1 mutations have accumulation of RANK in luminal progenitor cells and increased dual expression of oestrogen receptor and Ki67, proposed to be an early indicator of developing breast cancer [1,2]. In non-tumour tissue taken from cancer-containing breasts, we identified four distinct tissue subtypes: metabolic, immune, matrisome/EMT and non-coding enriched and found that patients in the metabolic subtype exhibited a significantly poorer prognosis [3]. Roman-Perez also identified distinct expression subtypes in “normal” tissue adjacent to tumour, overlapping with ours, that associated with patient outcome [4].
The hypothesis of this study is that molecular alterations – at DNA, RNA and protein level – precede morphological alterations in the development of breast cancer. Identification of these changes would allow improved risk stratification and guide screening and prevention programmes.
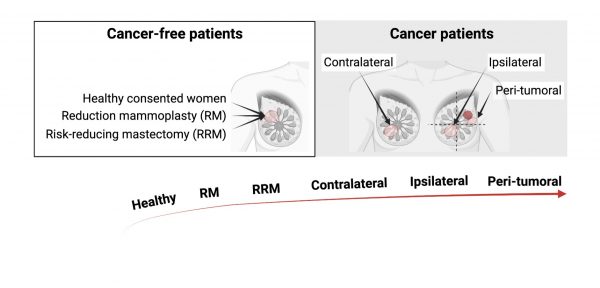
To address this, we will analyse the genomic, transcriptomic and proteomic profile of histologically normal breast tissue from well-defined patient cohorts across a spectrum of breast cancer risk. We have unique access to tissues and cells through the Breast Cancer Now Tissue Bank (BCNTB):
- Cosmetic reduction mammoplasty tissue with no known GL mutation (low-risk; n=50)
- RRM tissue from patients with known GL BRCA1/2 mutation (mod-high risk; n=80)
- Tissue adjacent to or distant from (including contralateral) breast cancer in patients with or without BRCA1/2 mutation (high-risk; n=100)
For some of these cases, including BRCA1/2 samples, we already have collected digitized images and using deep learning (DL)- methods, can capture epithelial, stroma and adipocytes which show alterations in normal tissue of these women.
We will build on this to:
- Carry out shallow whole genome sequencing for copy number alterations, RNAseq and phospho-proteomics;
- Perform an integrated analysis of omic data using an adapted pipeline from Sivapalan et al. [5] and Gadelata et al. [3] to define signatures of low- and high-risk;
- Validate these signatures in independent samples using a combination of spatial transcriptomics, proteomics and targeted NGS;
- In vitro validation of functional significance of novel hits using primary isolated cell populations (available through BCNTB), using knock-down and over-expression approaches.
This project provides a rare and exciting opportunity to gain experience at the interface of pathology, computational science, big data and functional studies in breast cancer.
Candidate background
A background in molecular biology and some experience of bioinformatics are desirable but not essential.
Potential Research Placements
- Pedro Cutillis, Barts Cancer Institute, Queen Mary University of London
- Iain Goulding, Breast Cancer Now Primary Cell Bank
- Anita Grigoriadis, Spatial Transcriptomic Facility, King’s College London
