Post-transcriptional regulation of cytotoxic T cell function in tumours
Primary supervisor: Richard Jenner, UCL
Secondary supervisor: Diu Nguyen, Queen Mary University of London
Project
Cancer immunotherapy approaches that seek to boost T cell function have been successful in providing benefits to a subset of patients. However, it is becoming clear that a large proportion of tumours remain resistant. A better understanding of how T cell effector function is regulated in tumours is critical for developing improved therapies.
We have previously defined transcriptional regulatory mechanisms underlying the acquisition of cytotoxic CD4+ T cell activity in tumours [1]. There is increasing evidence that T cell effector function is also tightly controlled at the post-transcriptional level by RNA binding proteins (RBPs) [2,3,4].
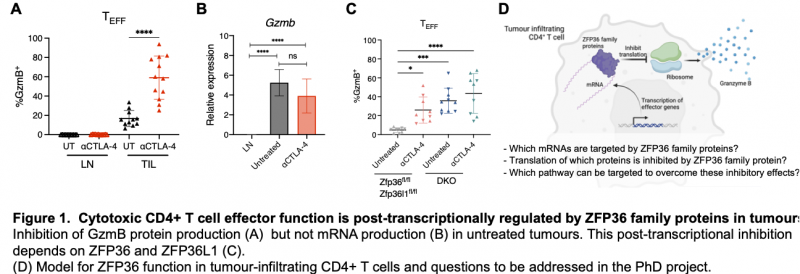
We have recently found that CD4+ T cells exist in an inactive but poised state in untreated tumours [5]. CD4+ T cells contain mRNA encoding the key cytotoxic effector Granzyme B (GzmB) but no protein (Fig. 1A-B). Treatment of tumours with anti-CTLA4, which depletes suppressive Tregs, relieves the block to GzmB protein production and triggers tumour regression. Thus, CD4+ effector T cell function is restrained in tumours by a post-transcriptional regulatory checkpoint. Understanding the mechanisms responsible could provide new targets to enhancer anti-tumour immune responses.
Using mouse genetic knockouts, we have discovered that the RBPs ZFP36 and ZFP36L1 block cytotoxic CD4+ T cell effector function in tumours (Fig. 1C). The aim of the PhD project is to identify the mechanisms through which these proteins mediate their repressive effects and assess their potential as new targets for therapy (Fig. 1D).
Identifying the transcripts targeted by RBPs in small cell populations in vivo is challenging. To address this, the student will harness the HyperTRIBE method, in which the RBP of interest is fused to the RNA editing enzyme ADAR, allowing target mRNAs to be identified by high-throughput sequencing. The student will transduce murine CD4+ T cells with inducible ADAR-tagged ZFP36 and ZFP36L1 constructs and transfer the modified cells into mouse tumour models. They will then harvest effector T cells from untreated and anti-CTLA4-treated tumours and identify the RNA editing events that mark binding by the ADAR-fused ZFP36 proteins.
Subsequently, the ZFP36 and ZFP36L1 mRNA binding events that are associated with repression of protein production will be identified using proteomics analysis of tumour-infiltrating CD4+ T cells from knockout vs wild-type mice. Finally, the contribution made by repression of specific pathways by ZFP36 and ZFP36L1 on T cell effector function will be tested by manipulation of the pathways using blocking antibodies, small molecules or knockout of key pathway genes (either using available mouse lines or by transfer of CRISPR-Cas9-modified T cells).
Summary: This thesis project will reveal the mechanisms through which post-transcriptional regulators inhibit effector T cell function in tumours and thus provide new therapeutic opportunities to enhance anti-tumour responses in patients.
Candidate background
The project would suit someone interested in applying cutting-edge genomics and proteomics techniques to cancer immunology. The candidate will have an undergraduate degree in immunology or molecular biology and will ideally have experience of cell culture, flow cytometry and PCR. The student should be interested in splitting their time between wet and dry lab environments.
Potential Research Placements
- Diu Nguyen, Barts Cancer Institute, Queen Mary University of London
- Sergio Quezada, Cancer Institute, UCL
- Silvia Surinova, Cancer Institute, UCL
References
- Sledzinska, A., Vila de Mucha, M., Bergerhoff, K., Hotblack, A., Demane, D.F., Ghorani, E., Akarca, A.U., Marzolini, M.A.V., Solomon, I., Vargas, F.A., et al. (2020). Regulatory T Cells Restrain Interleukin-2- and Blimp-1-Dependent Acquisition of Cytotoxic Function by CD4(+) T Cells. Immunity 52, 151-166
- Jurgens, A.P., Popovic, B., and Wolkers, M.C. (2021). T cells at work: How post-transcriptional mechanisms control T cell homeostasis and activation. Eur J Immunol 51, 2178-2187.
- Zhu, W.S., Wheeler, B.D., and Ansel, K.M. (2023). RNA circuits and RNA-binding proteins in T cells. Trends Immunol 44, 792-806.
- Petkau, G., Mitchell, T.J., Chakraborty, K., Bell, S.E., V, D.A., Matheson, L., Turner, D.J., Saveliev, A., Gizlenci, O., Salerno, F., et al. (2022). The timing of differentiation and potency of CD8 effector function is set by RNA binding proteins. Nat Commun 13, 2274.
- Vila de Mucha M, Costoya C, Hlond J, Sledzinska A, Uddin I, Navarrete Sanchez M, Shah M, Mastrokalos G, Karagianni D, Beattie G, et al. Post-transcriptional acquisition of cytotoxic activity from a poised state in tumor-infiltrating CD4+ T cells. Submitted.
