Leveraging the role of oncogenes and tumour suppressors genes in 1p-/1q+ myeloma
Primary supervisor: Kwee Yong, UCL
Advisor: Dinis Calado, Francis Crick Institute
Project
Myeloma, the second most common form of blood cancer, poses a significant challenge as it increases in incidence whilst remaining incurable [1]. Currently, our understanding of myeloma revolves around a model whereby a B-cell undergoes an initiating event and progresses through the different disease phases by acquiring secondary events. Events offering a competitive advantage, are selected for during the disease course, leading to both aggressive extramedullary, and treatment-resistant forms of the disease [2]. The acquisition of secondary events affecting both the short and long arm of chromosome 1 together are among the most common in these settings having been shown to increase both during disease progression and relapse [3-4]. This is known to upregulate a set of oncogenes such as MDM4 and SETDB1 on 1q and downregulate tumour suppressors such as TENT5C and FAF1 on 1p [2]. However, it is unknown how the co-occurrence of loss of tumour suppressors and gain of oncogenes affect malignant plasma cells thus hindering the management of these cases.
Hypothesis: We hypothesise that deconvoluting the complex interplay between tumour suppressors and oncogenes on chromosome 1 both in vivo and in vitro will explain the aggressive disease phenotype and unlock better therapeutic strategie
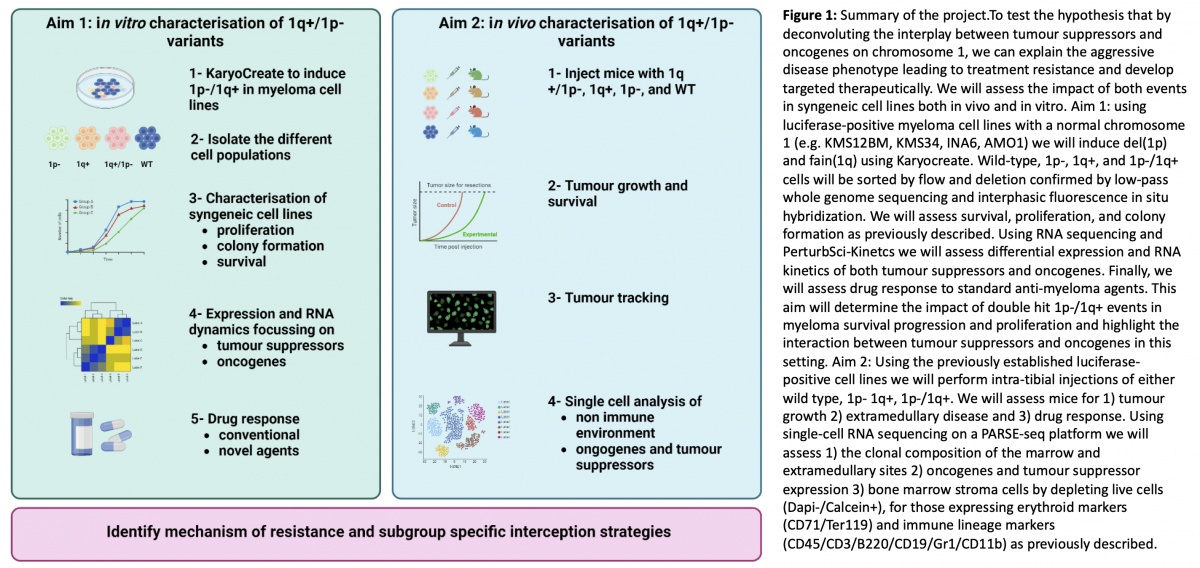
Aims and Objectives (Figure 1):
To test this hypothesis, the aims of this project are twofold:
- Aim 1: Assessing the impact of the co-occurrence of 1p-/1q+ on oncogenes and tumour suppressors in vitro. Using luciferase-positive myeloma cell lines with a normal chromosome 1, we will induce a 1p- and 1q+ using Karyocreate [5]. Cells will be sorted and assessed for survival, proliferation, and colony formation as previously described. Using RNA-sequencing and PerturbSci-Kinetics we will assess differential expression and RNA kinetics of transcription factors and tumour suppressors. Finally, we will assess drug response to standard anti-myeloma agents. This aim will 1) determine the impact of double-hit 1p-/1q+on cell survival, progression, and proliferation 2) establish the role of specific oncogenes and tumour suppressor gene interaction and 3) give mechanistic insight into this aggressive disease phenotype.
- Aim 2: Assessing the impact of the co-occurrence of 1p-/1q+ in vivo. Using the previously established luciferase-positive cell lines we will perform intra-tibial injections of either wild type, 1p-, 1q+ or 1p-/1q+ populations in NSG mice. We will assess mice for 1) tumour growth 2) extramedullary disease formation and 3) survival. Moreover, we will assess at the single-cell level 1) the clonal composition and evolution of the marrow and extramedullary sites and 2) the impact of these double-hit 1p-/1q+ cells on the bone-marrow stroma. This work will identify the impact of this double-hit event on 1) extramedullary disease 2) clonal evolution and 3) the non-immune microenvironment.
These objectives aim to shed light on the functional consequences of 1p-/1q+ alterations, and the role of specific tumour suppressors and oncogenic interaction providing insights into the mechanisms driving progression from precursors to relapsed and refractory disease.
Potential Applications and Benefits:
This research will uncover vulnerabilities in cells harbouring 1p-/1q+, paving the way for targeted therapeutic interventions in myeloma and potentially other cancers featuring similar chromosomal events.
References
- Kazandjian D. Multiple myeloma epidemiology and survival: A unique malignancy. Semin Oncol. 2016;43(6):676-681.
- Boyle EM, Davies FE. From little subclones grow mighty oaks. Nat Rev Clin Oncol 2023; 20: 141-142.
- Boyle EM*, Blaney P*, Stoeckle JH*, Wang Y, Ghamlouch H, Choi J, Braunstein M, Kaminetzsky D, Montes L, Corre J, et al Multiomic Mapping of Somatic Copy Number Abnormalities and Structural Variation to the Chromatin Structure of Chromosome 1 Highlights Multiple Recurrent Regions and Novel Disease Drivers, Clin Cancer Res Off J Am Assoc Cancer Res 2023, in press
- Boyle EM, Deshpande S, Ashby C, Tytarenko RG, Wang H, Wang Y et al. The Molecular Make Up of Smoldering Myeloma Highlights the Evolutionary Pathways Leading to Multiple Myeloma. Nat Commun 12, 293 (2021)
- Bosco N, Goldberg A, Zhao X, et al. KaryoCreate: A CRISPR-based technology to study chromosome-specific aneuploidy by targeting human centromeres. Cell. 2023;186(9):1985-2001.e19.
