ecDNA in circulating tumour DNA
Primary supervisor: Mariam Jamal-Hanjani, UCL
Secondary supervisor: Nnenna Kanu, UCL
Project
Extrachromosomal DNA (ecDNA) are circular genomic structures that can result in ultra-high copy number amplification and are associated with poor clinical outcome across multiple tumour types (1). ecDNA lack centromeres, consequently resulting in random segregation during cell division and fuelling intra-tumour heterogeneity. Currently, understanding of how ecDNAs form, how they can be monitored, and how they might be effectively targeted across different cancer types is lacking. Circulating tumour DNA (ctDNA) offers a window to study the evolving tumour genome, including structural variation, in a non-invasive manner, however, the study of ctDNA-based metrics as cancer biomarkers is in its infancy. The objective of this project is to develop ctDNA-based approaches to detect and monitor ecDNA, in order to facilitate biomarker and treatment development across cancer types.
This project will be carried out across two tumour types, non-small cell lung (NSCLC) (2) and breast (NCT03077776) cancers. These longitudinal studies have existing detailed clinical data integrated with state-of-the-art multi-omics. Importantly for this project, whole-genome sequencing (WGS) has already been carried out on a significant subset of patients, and longitudinal plasma samples collected. Data from both TRACERx programmes will be used to explore the utility of ctDNA for the identification and study of ecDNA during tumour evolution. Whilst the presence of this important biomarker has been described within solid tumour samples collected primarily at the time of surgical resection, the manner in which it accrues and fluctuates over time within patients – of particular interest given its esoteric pattern of molecular inheritance – remains relatively unstudied.
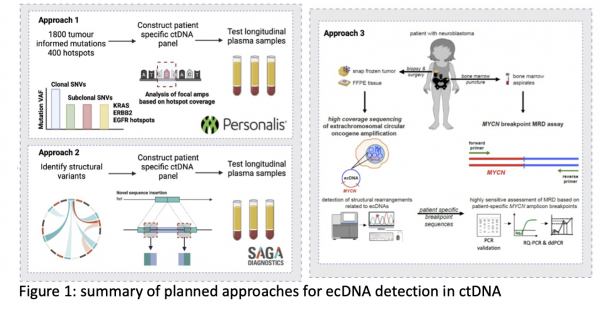
The breast analysis will benefit from an existing academic-commercial collaboration with SAGA Diagnostics, which aims to design personalised patient-specific fingerprints of structural variants (SVs) identified using tumour WGS for an ultrasensitive digital polymerase chain reaction (dPCR) assay performed in blood. The novelty of this work will be in establishing the stability of SVs (especially ecDNA), by leveraging multi-region biopsies of primary tumours, to track tumour evolutionary dynamics within ctDNA. For a subset of patients that demonstrated relapse following standard treatment, this data will afford us the unprecedented opportunity to interrogate ecDNA in a longitudinal setting as a predictor of adverse clinical outcomes. The NSCLC analysis will also benefit from an existing academic-commercial collaboration with Personalis and existing tumour-informed ctDNA data from which ecDNA detection may be feasible. Importantly, this work will benefit from the expertise and collaboration within the eDyNAmiC Cancer Grand Challenge team which we are part of and lead on the ctDNA analyses.
The main aims of this project are:
- Develop techniques for tumour-specific ecDNA detection in ctDNA (Figure 1)
- Identify limits of detection for ecDNA in ctDNA (e.g. copy number gain), differences between NSCLC and breast and clinical variables, e.g. stage and histology
- Investigate the dynamics of ecDNA evolution using longitudinal ctDNA
- Determine whether ecDNA in ctDNA can act as a surrogate measure of minimal residual disease (MRD)
- Investigate relationship between ecDNA in blood and clinical outcomes
- Orthogonal validation of a subset of ecDNA identified on WGS and ctDNA using matched tumour samples and FISH.
References
- Kim H et al. Extrachromosomal DNA is associated with oncogene amplification and poor outcome across multiple cancers. Nat Genet 52, 891-897 (2020)
- Jamal-Hanjani, M. et al. Tracking the Evolution of Non-Small-Cell Lung Cancer. N Engl J Med 376, 2109-2121 (2017).
