Investigating the role of DNA Polymerase Epsilon in sensitization to PARP inhibitors
Primary supervisor: Roberto Bellelli, Queen Mary University of London
Secondary supervisor: David Michod, UCL
Project
Genomic instability is a hallmark of cancer and understanding its nature has provided avenues for patient-tailored therapies. The most prominent example has been the successful use of PARP (Poly-ADP Ribose Polymerase) inhibitors (PARPi) in BRCA1/BRCA2 (Breast Cancer susceptibility 1/2) mutant cancers [1]. We have recently discovered that loss of the POLE3-POLE4 subunits of DNA Polymerase Epsilon (Polε) sensitizes cancer cells to PARPi, by unleashing replicative gap accumulation, and bypasses common mechanisms of resistance to these compounds [2]. Aim of this project is to understand the mechanism responsible for these phenomena to ultimately exploit Polε as novel therapeutic target. The project will combine approaches of biochemistry, structural biology and cell biology, available within CRUK City of London, to accomplish two aims:
Aim 1. Reconstitution of DNA replication and histone chaperoning by Polε.
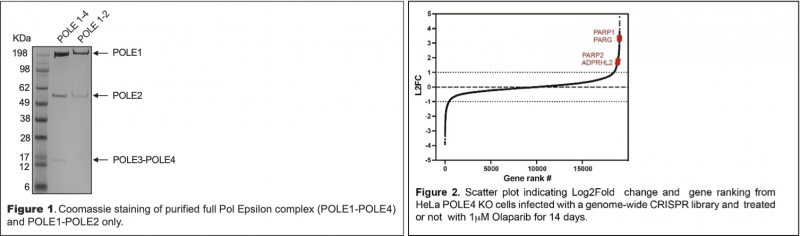
We previously discovered that the POLE3-POLE4 sub-complex binds double-stranded DNA [3]. This activity may promote Polε processivity, thus preventing replicative gap accumulation and sensitization to PARPi [2]. To address this hypothesis, we will investigate in vitro Polε-dependent DNA synthesis in the presence and absence of POLE3-POLE4 (Fig. 1). POLE3-POLE4 can also bind histone H3-H4 and participate in replication coupled-nucleosome assembly [3]. To determine the relevance of this activity in replicative gap formation and sensitization to PARPi, we will investigate the interaction with H3-H4 within full Polε and identify the domains involved by structural biology approaches (including CRYO-EM). This will allow the generation of separation-of-function mutants of Polε.
Aim 2. Understanding the mechanism of replicative gap formation upon treatment with PARPi
To understand why and how loss of POLE3-POLE4 leads to sensitivity to PARPi we have performed a genome-wide CRISPR survival screen in POLE4 KO cells treated with PARPi (Olaparib). Importantly, PARP1 and PARP2 scored among top hits in the resistance arm of the screen, pointing to “trapping” of PARP1-PARP2 being crucial for sensitization to these compounds (Fig. 2). Which DNA lesion(s) triggers PARP1-PARP2 chromatin binding, and consequent trapping, in POLE3-POLE4 KO cells, remains however to be determined. Additionally, whether inhibition of PARP1-PARP2 enzymatic activity is required to sensitize POLE3-POLE4 KO cells remains to be clarified. To address these questions, we will generate PARP1-PARP2 single and double KO cells in a WT and POLE3-POLE4 KO background and investigate sensitivity to PARPi and replication dynamics/replicative gap formation by DNA fiber assay-based approaches, well established in my laboratory [2]. Additionally, previous work has suggested that inhibition of fork reversal by PARPi has an important role in sensitization to these compounds [4]. Thus, we will also investigate the role of SMARCAL1 and RECQ1, crucial enzymes involved in this process, in mediating replicative gap formation and sensitization to PARPi, using similar approaches. Finally, we will employ eSPAN (enrichment of Proteins on Nascent DNA) a technique we recently set up with Dr Michod, to study the role played by the histone chaperone activity of εPol in genome stability and sensitization to PARPi [5].
Candidate background
This project would suit candidates with a background in biochemistry and cell biology and an interest in genome stability.
Potential Research Placements
- David Michod, Institute for Child Health, UCL
- Peter Cherepanov, Francis Crick Institute
- Nnenna Kanu, Cancer Institute, UCL
References
- Lord CJ, Ashworth A. PARP inhibitors: Synthetic lethality in the clinic. Science. 2017 Mar 17;355(6330):1152-1158.
- Hill BR, Ozgencil M, Buckley-Benbow L, Skingsley SLP, Tomlinson D, Eizmendi CO, Agnarelli A, Bellelli R. Loss of POLE3-POLE4 unleashes replicative gap accumulation upon treatment with PARP inhibitors. Cell Rep. 2024 May 28;43(5):114205.
- Bellelli R, Belan O, Pye VE, Clement C, Maslen SL, Skehel JM, Cherepanov P, Almouzni G, Boulton SJ. POLE3-POLE4 Is a Histone H3-H4 Chaperone that Maintains Chromatin Integrity during DNA Replication. Mol. Cell 72,112-126.
- Quinet A, Tirman S, Jackson J, Šviković S, Lemaçon D, Carvajal-Maldonado D, González-Acosta D, Vessoni AT, Cybulla E, Wood M, Tavis S, Batista LFZ, Méndez J, Sale JE, Vindigni A. PRIMPOL-Mediated Adaptive Response Suppresses Replication Fork Reversal in BRCA-Deficient Cells. Mol Cell. 2020 Feb 6;77(3):461-474.
- Li Z, Hua X, Serra-Cardona A, Xu X, Zhang Z. Efficient and strand-specific profiling of replicating chromatin with enrichment and sequencing of protein-associated nascent DNA in mammalian cells. Nat Protoc. 2021 May;16(5):2698-2721.
