Modelling the evolution and ecology of cancer resistance to develop new proton radiobiology insights
Primary supervisor: Maria Secrier, UCL
Secondary supervisor: Maria Hawkins, UCL
Advisor: Ginestra Bianconi, Queen Mary University of London
Project
Cancer is a highly plastic system where tumour cells continuously adapt to their niche while actively remodelling it in order to persist and grow within the tissue. This highly effective co-adaptation with the tumour microenvironment is what makes some cancers so successful in evading even the most commonly employed anti-cancer therapies, such as radiotherapy. Radiotherapy resistance is seen in nearly half of patients receiving radiotherapy for colorectal cancer and potentially varies depending on the mode of action (photon vs proton) [1]. In the cases where treatment resistance emerges, this is often driven by “persister cells”, which enter a slow cycling/arrested state during which they accumulate resistance mutations that lead to drug tolerance. The persister state is difficult to capture as it is generally short-lived, yet it is pervasive across all major therapies. Early evidence shows that persister cells co-opt the tumour microenvironment to persist and disseminate. Promptly targeting these resistant cells remaining after the first-line therapy is critical for preventing tumour local and distant recurrence, but we lack robust biomarkers to identify them. Furthermore, the effects of proton therapy versus X-ray on the emergence of resistance are poorly understood.
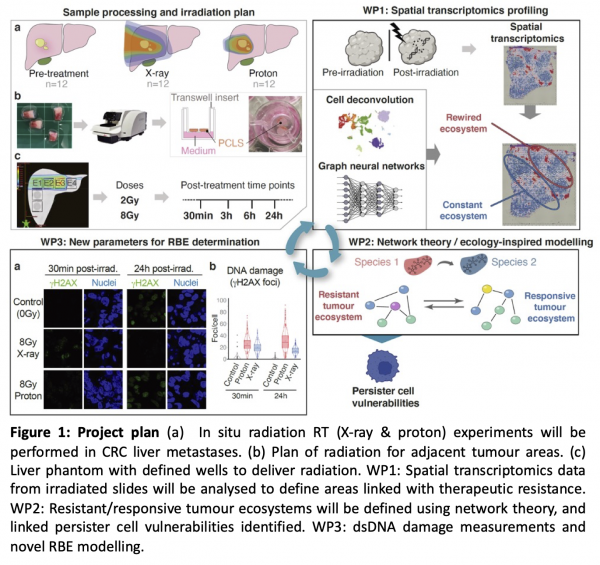
Recent work in the Secrier lab has uncovered a robust transcriptional signature capturing a rapid, short-lived tumour cell adaptation that enables drug tolerance via temporary arrest in the cell cycle, similar to a persister cell state [2]. We propose that our cell cycle arrest signature provides the ideal baseline to study persister cells and the resistant ecosystem they create within the tumour upon radiation. This project, jointly led by the Secrier (UCL), Hawkins (UCL) and Bianconi (QMUL) labs, will combine wet lab experiments, clinical knowledge, bioinformatics, AI, machine learning and network theory to test this hypothesis and study the effects of X-ray and proton radiotherapy in a controlled in situ setting (Figure 1). We will use spatial transcriptomics data from ex vivo irradiated human colorectal metastases at multiple time points before and after treatment to perform a multiscale characterisation of the evolution and environment of persister cells in the context of proton and photon-based radiotherapy. The underlying biological information will be used to identify new biomarkers for radiation resistance, develop a novel method to understand proton biological effectiveness and unlock new radiation-drug combination possibilities. We propose a unique setup combining human cancer precision cut slices, spatial biology and advanced analytics. This project will create new cross-disciplinary methods inspired from computer science and mathematics to deliver a new understanding of early radiation resistance and a novel complex in vitro model for proton relative biological effectiveness.
References
- O’Cathail SM, Smith T, Owens R, Zeniou A, Tsang Y, Holyoake DLP, Murray L, Harrison M, Hawkins MA. Superior outcomes of nodal metastases compared to visceral sites in oligometastatic colorectal cancer treated with stereotactic ablative radiotherapy. Radiother Oncol 2020
- Wiecek AJ, Cutty SJ, Kornai D, Parreno-Centeno M, Gourmet LE, Tagliazucchi GM, Jacobson DH, Zhang P, Xiong L, Bond GL, Barr AR, Secrier M. Genomic hallmarks and therapeutic implications of G0 cell cycle arrest in can. Genome Biol 2023
- Malagoli Tagliazucchi G, Wiecek AJ, Withnell E, Secrier M. Genomic and microenvironmental heterogeneity shaping epithelial-to-mesenchymal trajectories in cancer. Nat Commun 2023
- Withnell E, Secrier M. SpottedPy quantifies relationships between spatial transcriptomic hotspots and uncovers new environmental cues of epithelial-mesenchymal plasticity in cancer. bioRxiv 2023
- Millán A.P., Torres JJ, Bianconi G. Explosive Higher-Order Kuramoto Dynamics on Simplicial Complexes. Phys Rev Letters 2017
