Human Tumour Microenvironment Models
The aim of our research is to establish a multicellular tumour cell-agnostic iPSC-based tumour microenvironment (TME) model that incorporates immune elements of the TME and is applicable to studying a range of solid cancers. In particular, we are interested in understanding malignant cell-tumour associated macrophage (TAM) crosstalk and how different tumour cell types influence myeloid cell phenotype.
By using an engineered collagen hydrogel-based culture approach, iPS cells differentiated into macrophages and primary fibroblasts, we are validating the system on commercially available cell lines, as well as malignant cell lines established in our lab and provided by collaborators from CRUK CoL network.
We envisage this model will provide a standardised platform for developing new biological therapies targeting myeloid cells in the TME.
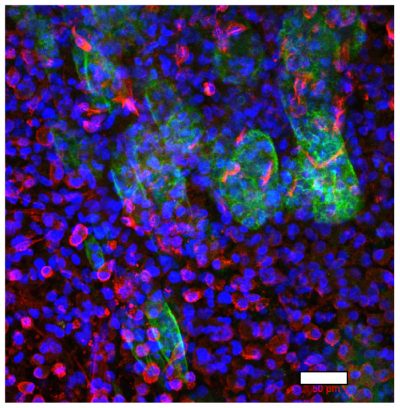
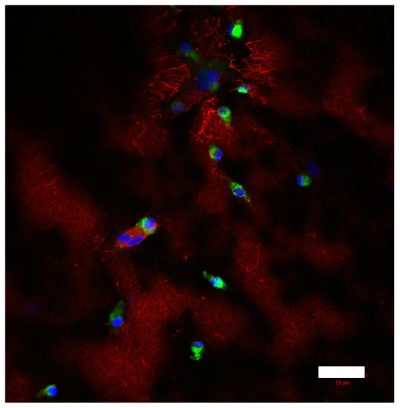
A multicellular colorectal cancer TME model with primary fibroblasts and iPS-derived macrophages
Left, colorectal cancer cells (green) and primary fibroblasts (red) at the border between malignant cells and stroma; right, CD68-positive iPS-derived macrophages (green) and fibronectin (red), deposited in stromal compartment of the model. Nuclei are stained in blue. Scale bar=50µm
Contact
Academic Lead
Ongoing projects/collaborations
Collaborators: Subhankar Mukhopadhyay (KCL)
Immortalised macrophage-like cell lines and peripheral blood-derived macrophages are currently the standard source of myeloid cells in in vitro cancer research. Nevertheless, accumulation of genetic aberrations and functional defects as well as high level of variability and difficulty for genetic manipulation limits their applicability. Macrophages derived from iPS cells have similar phenotypes to that of primary macrophages, are amenable to genetic manipulation and provide a standardised source for derivation of myeloid cells.
Supported by our collaborator we established a feeder-dependent method for growing iPSC-derived macrophages (iPSCDM). As a more time-efficient and less labour-intensive alternative, we have now established a feeder-independent protocol for iPSCDM and iPSCDM have already being used in other projects.
We aim at providing the protocol for iPSCDM as a service and SOP to be used in ongoing projects of our lab and to be made available to potential collaborators across CRUK CoL Centre. Please get in touch with us to discuss further details.
Collaborators: Umber Cheema (UCL)
Tumour associated macrophages (TAMs) are among the main immune components in the TME of different solid cancers. Accumulating data from studies looking at the prognostic associations of immune infiltrates show that high densities of TAMs are indicative of poor prognosis and overall patient survival. Targeting the pro-tumour behaviour of TAMs, such as inhibiting monocyte recruitment, depletion of TAMs, and functional/phenotypic reprogramming can be an alternative immunotherapy approach.
The objective of this study focuses on gaining a better understanding of TAM interactions with malignant cells and other TME elements. We are using a bioengineered TME model (3D raft culture), established by our collaborators and we increase its complexity by co-culturing of malignant cells with stromal non-immune cells (fibroblasts) and macrophages derived from peripheral blood and iPS cells. We are currently developing downstream endpoint quantitative analyses on the model (IF, IHC, Flow cytometry).
So far, we have used colorectal cancer, clear cell ovarian cancer and high grade serous ovarian carcinoma cell lines and we are open to new collaborations.
Additional information
- iPSC-derived macrophage protocol: https://www.jove.com/t/61038/production-characterization-human-macrophages-from-pluripotent-stem
- 3D raft culture model: https://www.nature.com/articles/srep44045